Summary
|
The COVID-19 pandemic has emphasized the need for a more robust disease surveillance infrastructure. However, the development of such an infrastructure runs into a double-bind. Assurances of privacy require adversarial testing against realistic systems. Likewise, it is difficult to build realistic systems without access to data. To get around this problem, we created synthetic datasets that reflect a realistic disease outbreak. This data was used as a component in the US-UK Prize Challenge on Privacy-Enhancing Technologies. In the challenge, participants were tasked with creating personalized risk forecasts of infection in a privacy-preserving manner. The challenge was put on by the U.K.’s Center for Data Ethics and Innovation (CDEI) and Innovate UK, as well as by the U.S. National Institutes of Standards and Technology (NIST), and the National Science Foundation (NSF) in cooperation with the White House Office of Science and Technology Policy (OSTP). The winners of the US and UK challenges were announced on March 30, at the Summit for Democracy. The winning teams can be found here, and profiles of the teams can be found here. The US-UK privacy-enhancing technologies (PETs) prize challenges were followed by a roundtable workshop. The workshop was a half-day meeting that brought together researchers, technologists, policy analysts, data regulators, and data ethicists to discuss ideas around PETs for pandemic preparedness and response, and healthcare data. The roundtable report with comments from participants after the roundtable can be found here. |
 |
What is in this dataset?
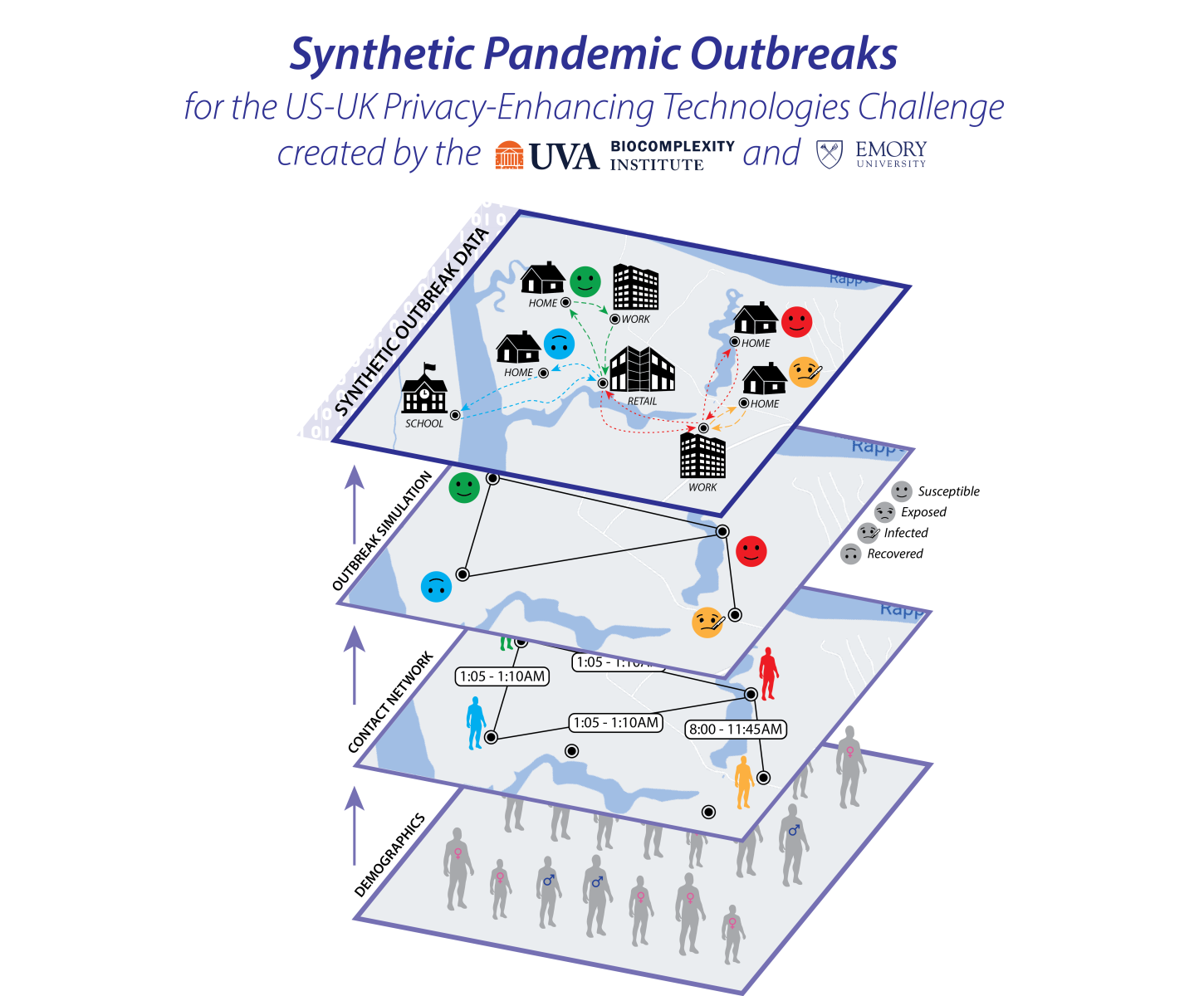
This dataset is synthetic data, sometimes called a virtual population or digital twin. It is a statistically accurate representation of a real population's demographics, activities, and social contacts, but does not contain any individual person’s information. This dataset consists of 3 components:
- A synthetic population representing a demographically realistic population, including synthetic activities for each individual denoting the different locations they visited, when they visited, and the duration of that visit;
- A contact network derived from these activities; and
- A set of disease states, the result of simulating the effects of a COVID-like disease within this population.
There are two datasets available. One is a representation of the state of Virginia (~7.7 million individuals) and the other is a representation of the United Kingdom (~62 million individuals).
Where did this data come from?
This data was synthesized. We combined multiple sources of publicly and commercially available data to produce data that statistically resembles what the real data would be if it were available. No entry in this dataset corresponds to any real person’s data. For more details about how this data was produced, please see our technical report.
How accurate is this data?
To synthesize this data we use high-level demographic census-type data. The synthetic population, activities, and contact network are constructed to satisfy to the greatest extent possible observed marginal distributions but are limited to the available data. As such the synthetic data will resemble the data that can be collected from the actual population, but will not be an accurate representation of any single individual. We do not model day-to-day variation in activities - all synthetic individuals do the same activities every day, though activity sequences may differ between individuals, and they do not change their behavior in response to disease status. The simulation of a COVID-like disease is also a simplification. There is a single contagion process, and we do not model differing immune statuses or vaccinations.
What was this data originally used for?
The data was used as part of the joint US-UK Challenge on Privacy-Enhancing Technologies. In the competition, participants were asked to build privacy-preserving learning systems, which were evaluated both on how well they performed and also on how well they protected privacy.
Participants were charged with using our data to try to forecast the risk of infection for each person in the synthetic population, given observations of infection state at some time. An effective private solution to this task would be able to provide a person with a forecast of how likely they are to be infected over the next week, similar to a pollen count or air quality report.
The US competition was sponsored by NIST and NSF with collaboration from OSTP, and The UK competition was conducted by Innovate UK and the Centre for Data Ethics and Innovation. DrivenData provided hosting and implementation for both the US challenge and the UK challenge, and was part of the set of prize challenges for Democracy-Affirming technologies announced at the Summit for Democracy. Learn about how the US and UK winners approached the challenge.
Why not use real data?
Privacy issues make the creation or sharing of real data with equivalent granularity or size infeasible. Real mobility data linked to specific individuals, joined with COVID infection status would be incredibly sensitive data. Even if such data existed, access requirements would make conducting a challenge using this data highly burdensome. Furthermore, our dataset is much larger and more comprehensive than one might be able to collect via sampling. The synthetic data approach we use allows more data that is more easily shared. In fact, synthetic data is itself an example of a privacy-enhancing technology.
Where can I get this data, and how should I cite it?
The data is available here. This is a permanent link to the data. Find the data files on this page by accessing the link noted next to “Other Location for Dataset” (directly under the citation at the top or under the Metadata tab).
Please cite this data as follows:
Acknowledgements
This work is based on the dataset and methods developed under NSF RAPID: COVID-19 Response Support: Building Synthetic Multi-scale Networks, NSF Expeditions in Computing, and the University of Virginia Strategic Investment Fund. The authors would like to thank members of the Biocomplexity COVID-19 Response Team, Network Systems Science and Advanced Computing (NSSAC) Division, University of Virginia. Special thanks to Erin Raymond, Lily Li, Golda Barrow, and Kristy Hall for their incredible and timely support. We also thank colleagues at NSF (James Joshi), NIST (Diane Ridgeway, Naomi Lefkovitz, Jim Horan, Joe Near), NSF/OSTP (Tess DeBlanc-Knowles), Driven Data (Jay Qi and Christine Chung) CDC (Matt Biggerstaff), and UK (Mark Durkee, David Buckley).
This work was partially supported by the National Institutes of Health (NIH) Grant R01GM109718, VDH Grant PV-BII VDH COVID-19 Modeling Program VDH-21-501-0135, NSF Grant No.:OAC-1916805, NSF Expeditions in Computing Grant CCF-1918656, CCF-1917819, NSF RAPID CNS-2028004, NSF RAPID OAC-2027541, NSF PREPARE CNS-2041952, US Centers for Disease Control and Prevention 75D30119C05935, NSF XSEDE TG-BIO210084 and NSF Prepare grant. Any opinions, findings, conclusions, or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies. This work used resources, services, and support from the COVID-19 HPC Consortium, a private-public effort uniting government, industry, and academic leaders who are volunteering free compute time and resources in support of COVID-19 research.



